Table of Contents
Introduction
One step in the machine learning process is feature engineering and within that step the selection of relevant features can help to reduce the complexity of the model. Several methods are described in Feature Engineering and Selection: A Practical Approach for Predictive Models by Max Kuhn and Kjell Johnson.
An example of non-greedy wrapper methods is Genetic Algorithm (GA). GA is an optimisation tool based on evolutionary biology and is able to locate the optimum or near optimum solutions for complex functions. Before we look at how the algorithm works and how it can be performed with the programming language R lets look at the naming convention:
- Genes => Features
- Chromosome => Feature Vector
- Population => Set of Feature Vectors for one Iteration
- Generation => One optimisation iteration
GA uses two methods to change the selection of features and to avoid local minima while optimising the feature set, those are:
- Crossover
- Mutation
The two mechanisms are show in the animation below. The animation is produced with the gganimate package. The status of the genes on the left-hand side of the crossover point are exchanged between chromosomes, while in right-hand side chromosomes change their status only via mutation. The probability of both mechanisms are parameters of the GA.
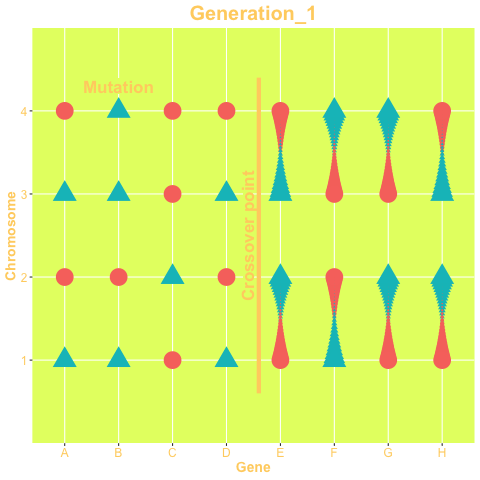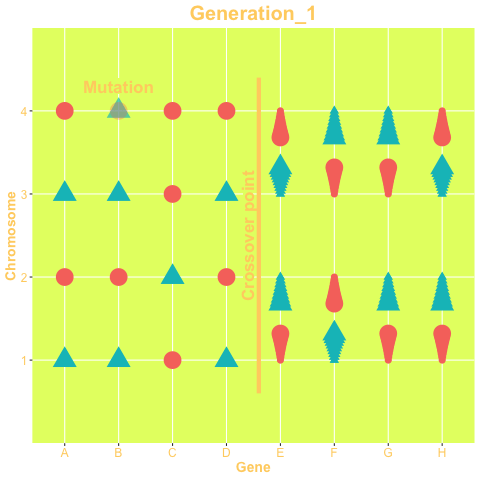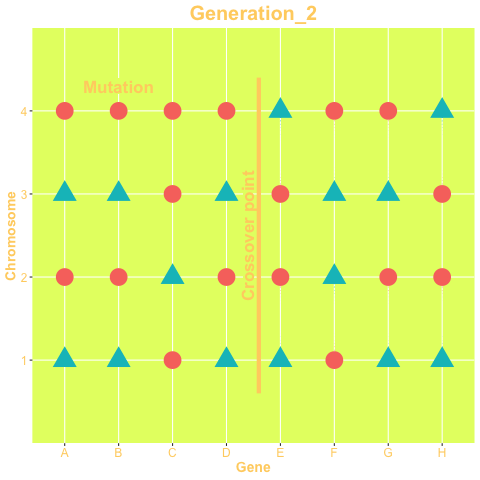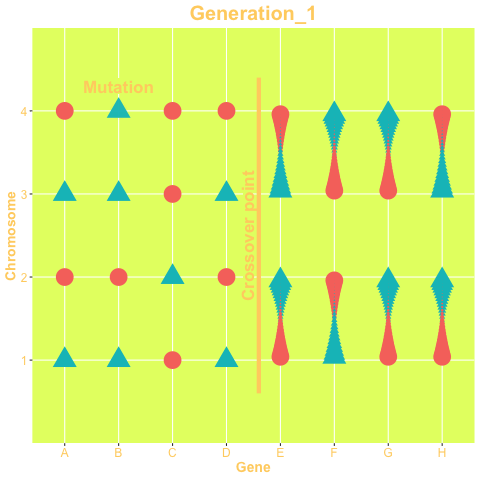
Using GA in R with caret
Using the gafs function of Max Kuhn’s caret R package makes the feature selection with GA straight forward as seen in the following code snippet. Note, since the operation is rather computational intensive it is advisable to parallelise the execution, example is for Mac OS.
cores=detectCores()
cl <- makeCluster(cores[1]-1) #not to overload your computer
registerDoParallel(cl)
ga_ctrl <- gafsControl(functions = rfGA,
method = "repeatedcv",
repeats = 5,
allowParallel = TRUE)
set.seed(10)
rf_ga <- gafs(x = x, y = simData$target,
popSize = 5,
iters = 10,
gafsControl = ga_ctrl)
stopCluster(cl)Results
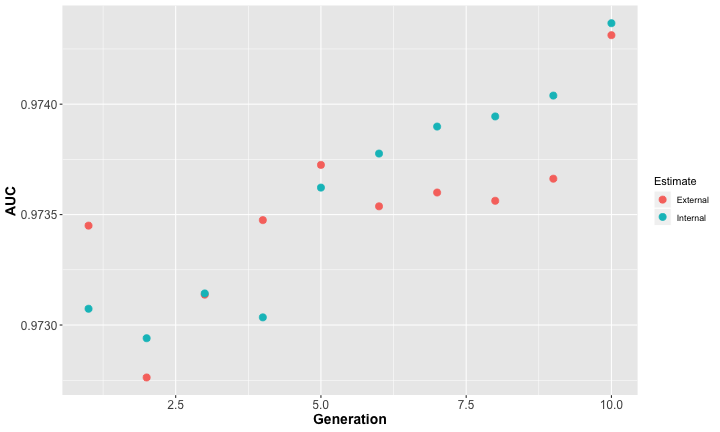
The improvement of the target metric vs generations can be plotted to judge how much the reduced feature set helps and if further generations are needed. However, even a slightly worse performance along with a significant reduction in model complexity might be a useful features set. In the following graph an improvement of the metric can clearly be seen. Explanations of the expressions external, internal in the graph can be found here
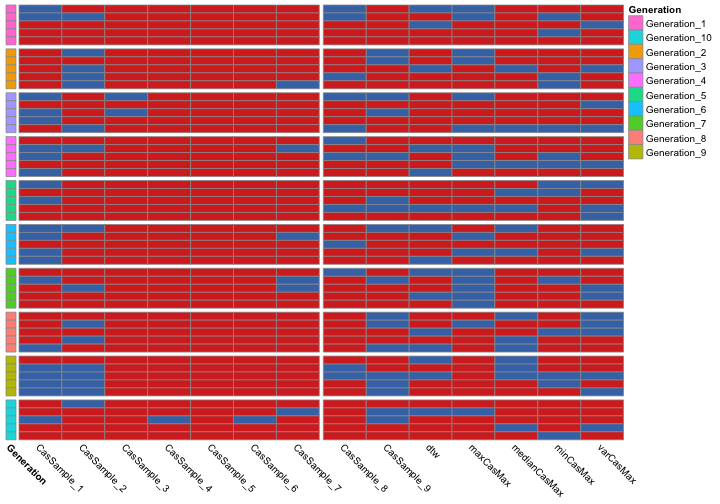
The combinations of features being tested are shown in the graph below. The GA parameters are:
- Maximum generations: 10
- Population per generation: 5
- Crossover probability: 0.8
- Mutation probability: 0.1
- Elitism: 0
Generations are marked by coloured boxes on the left-hand side of the graph. The names of the features on the x-axis are cropped on purpose.
- Red => Feature selected in chromosome
- Blue => Feature deselected in chromosome
Summary
A short intro to feature selection using GA alongside a R example which is hopefully enough for you to start experimenting to find a good feature set. However, be cautious, a good feature set for one model might not be a good one for another, i.e. a good feature set for random forest might not work for SVM.
I would be very happy to get some feedback about your experience or questions.
Happy ML and R coding